D I G E S T S
doi:10.1111/evo.13873
Digest: Lab versus nature: Disease resistance evolution differs between environments ∗
Kim-Sara Wagner1,2 and Jelena Rajkov1
1GEOMAR Helmholtz Centre for Ocean Research, Marine Evolutionary Ecology, D ¨usternbrooker Weg 20, 24105 Kiel, Germany
2E-mail: kwagner@geomar.de
Received September 8, 2019 Accepted October 18, 2019
Does disease resistance evolutionin vitroreflect resistance evolutionin vivo? Hernandez and Koskella conducted serial passage experiments of the plant pathogenic bacteriumPseudomonas syringaeand two lytic bacteriophages in high-nutrient medium (in vitro) and in a tomato plant (in vivo). High levels of bacterial resistance to phages evolvedin vitrobut notin vivo, suggesting that high costs and low benefits of resistance explain the observed pattern.
Viruses are the most abundant biological entities and harbor the greatest genetic diversity on the planet, and we are rapidly learning about their ecology and evolution in natural popu- lations. Bacteriophages, or viruses that specifically infect and replicate within prokaryotes, are an important killing agent of bacteria and, as such, act as a key selective force in bacterial communities.
One of the key questions in the field of disease ecology con- cerns the coevolution of pathogen virulence, and host resistance.
Host resistance strategies can reduce the probability of infection, limit pathogen replication within the host, or accelerate recovery through pathogen clearance. However, these resistance strategies often come at a cost: in bacteria, infection occurs via the adsorp- tion of the phage to specific surface receptors on the bacterial host.
To prevent adsorption, bacteria can either downregulate, block, or lose the receptor that, in turn, can impact their ability to take up certain nutrients, leading to a decreased competitive ability (van Houte et al. 2016). Fitness costs of resistance mutations therefore depend on the environment (Bohannan and Lenski 1997; Harrison et al. 2013). For example, mutations underlying phage resistance were shown to be more costly in terms of bacterial growth in plant leaves than in nutrient broth (Meaden et al. 2015). Resis- tance to phages commonly evolves when bacteria are culturedin
∗This article corresponds to Hernandez, C. A., and B. Koskella. 2019.
Phage resistance evolutionin vitrois not reflective ofin vivooutcome in a plant-bacteria-phage system. Evolution. https://doi.org/10.1111/evo.13833.
vitro, but few studies have investigated phage resistance evolution within natural bacterial hosts such as plants or animals (Oechslin 2018).
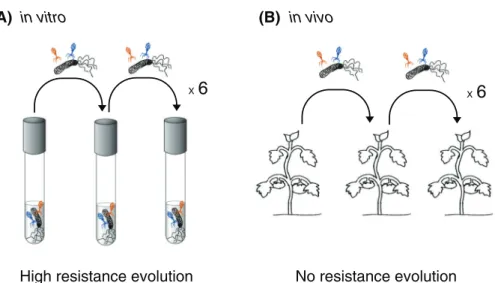
In this issue, Hernandez and Koskella (2019) use lytic phages and their bacterial host—the tomato plant pathogenPseudomonas syringae—to provide a direct comparison of phage resistance evo- lution between two distinct environments: a high-nutrient medium (in vitro; Fig. 1A) and a tomato plant (in vivo; Fig. 1B). In each environment, the authors conducted serial passage experiments, which enable the study of reciprocal virulence and resistance co- evolution through the propagation of pathogens under defined conditions and the comparison of the newly evolved pathogen characteristics to those of the ancestral state.
The authors conducted six serial passages of the bacteria with or without a lytic phage. In a second experiment, they further investigated (1) the effect of higher phage selection pressure using higher phage concentration, (2) the effect of another lytic phage, and (3) the effect of a “cocktail” of both phages simultaneously.
Whereas phage-selected resistance consistently evolved in high- nutrient medium, it was absent or negligible inside the tomato plant, in which the phage was replicating, but at much lower rates than in the medium.
To investigate the benefits of resistance evolved in vitro, the authors grew resistant mutants together with the sensitive bacteria, bothin vitroandin vivo. In the medium, in the presence of the phage, the resistant mutant bacteria grew faster than the non-resistant bacteria. Inside the plant host, however, the
2 5 4 0
C2019 The Author(s).EvolutionC2019 The Society for the Study of Evolution.
Evolution 73-12: 2540–2541
D I G E S T
No resistance evolution
X
6
High resistance evolution
X
6
(A) in vitro (B) in vivo
Figure 1. Serial passage experiments of the plant pathogenPseudomonas syringaeand two lytic bacteriophages conductedin vitro—in a test tube with high-nutrient medium (A)—andin vivo—in the leaf of a tomato plant that represents the bacterial natural host (B).
Bacteria became resistant to phages only in high-nutrient medium. Drawings by Alba Filella and Tatjana Liese.
phage-resistant mutants had no growth advantage. This suggests that higher cost and lower benefits of resistance evolution in the plant relative to the high-nutrient medium impede the evolution of resistance.
This study paves the way for future research and application in medicine and agriculture. After decades of excessive usage of antibiotics, the spread of multidrug-resistant diseases is a major global health issue that calls for novel therapeutic approaches.
Phage therapy, in which bacteriophages are used as antimicrobial agents to fight infections, has already shown promising results in eliminating cystic fibrosis strains ofPseudomonas aeruginosain selected clinical trials (Kortright et al. 2019). InP. aeruginosa, phage-resistant mutants emergedin vitrobut notin vivo, likely be- cause resistance mutations affected bacterial surface determinants important for infectivity (Oechslin et al. 2017). Phage therapy has the potential to circumvent fast resistance evolution, which is the main problem that has arisen with the usage of antibiotics. Fu- ture studies on phage therapy should focus on trying to increase the phage replication rate inside the host to eventually combine phage therapy with conventional antibiotics. It is thus time to move away from lab conditions to more natural environments for studying disease resistance evolution.
LITERATURE CITED
Bohannan, B. J. M., and R. E. Lenski. 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78:2303–
2315.
Harrison, E., A. L. Laine, M. Hietala, and M. A. Brockhurst. 2013. Rapidly fluctuating environments constrain coevolutionary arms races by imped- ing selective sweeps. Proc. R. Soc. B Biol. Sci. 280.
Hernandez, C. A., and B. Koskella. 2019. Phage resistance evolutionin vitro is not reflective ofin vivooutcome in a plant-bacteria-phage system.
Evolution. https://doi.org/10.1111/evo.13833
Kortright, K. E., B. K. Chan, J. L. Koff, and P. E. Turner. 2019. Phage therapy:
a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25:219–232.
Meaden, S., K. Paszkiewicz, and B. Koskella. 2015. The cost of phage resis- tance in a plant pathogenic bacterium is context-dependent. Evolution.
69:1321–1328.
Oechslin, F. 2018. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10:351.
Oechslin, F., P. Piccardi, S. Mancini, J. Gabard, P. Moreillon, J. M. Entenza, G. Resch, and Y. A. Que. 2017. Synergistic interaction between phage therapy and antibiotics clearsPseudomonas aeruginosainfection in en- docarditis and reduces virulence. J. Infect. Dis. 215:703–712.
van Houte, S., A. Buckling, and E. R. Westra. 2016. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 80:745–
763.
Associate Editor: K. Moore Handling Editor: T. Chapman
SUBMIT A DIGEST
Digests are short (500 word), news articles about selected original research included in the journal, written by students or postdocs. These digests are published online and linked to their corresponding original research articles. For instructions on Digests preparation and submission, please visit the following link: https://sites.duke.edu/evodigests.
EVOLUTION DECEMBER 2019 2 5 4 1