ContentslistsavailableatScienceDirect
Systematic and Applied Microbiology
j ou rn a l h om ep a ge :h t t p : / / w w w . e l s e v i e r . c o m / l o c a t e / s y a p m
Pseudooceanicola algae sp. nov., isolated from the marine macroalga Fucus spiralis, shows genomic and physiological adaptations for an algae-associated lifestyle
Laura A. Wolter
a,b,∗, Matthias Wietz
a,c, Lisa Ziesche
d, Sven Breider
a, Janina Leinberger
a, Anja Poehlein
e, Rolf Daniel
e, Stefan Schulz
d, Thorsten Brinkhoff
a,∗∗aInstituteforChemistryandBiologyoftheMarineEnvironment,Oldenburg,Germany
bJSTERATONomuraProject,FacultyofLifeandEnvironmentalSciences,Tsukuba,Japan
cAlfredWegenerInstituteHelmholtzCentreforPolarandMarineResearch,Bremerhaven,Germany
dInstituteofOrganicChemistry,TechnischeUniversitätBraunschweig,Germany
eInstituteofMicrobiologyandGenetics,GenomicandAppliedMicrobiology,andGöttingenGenomicsLaboratory,Germany
a rt i c l e i nf o
Articlehistory:
Received2September2020
Receivedinrevisedform9November2020 Accepted12November2020
Keywords:
Roseobactergroup Algae-associatedlifestyle Tidalflat
Comparativegenomics Secondarymetabolites Habitatadaptations
a b s t ra c t
Thegenus PseudooceanicolafromthealphaproteobacterialRoseobactergroup currentlyincludesten validatedspecies.WehereindescribestrainLw-13eT,thefirstPseudooceanicolaspeciesfrommarine macroalgae,isolatedfromthebrownalgaFucusspiralisabundantatEuropeanandNorthAmericancoasts.
PhysiologicalandpangenomeanalysesofLw-13eT showedcorrespondingadaptivefeatures.Adapta- tionstothetidalenvironmentincludeabroadsalinitytolerance,degradationofmacroalgae-derived substrates(mannitol,mannose,proline),andresistancetoseveralantibioticsandheavymetals.Notably, Lw-13eTcandegradeoligomericalginateviaPL15alginatelyaseencodedinapolysaccharideutilization locus(PUL),rarelydescribedforroseobacterstodate.PlasmidlocalizationofthePULstrengthensthe importanceofmobilegeneticelementsforevolutionaryadaptationswithintheRoseobactergroup.PL15 homologswereprimarilydetectedinmarineplant-associatedmetagenomesfromcoastalenvironments butnotintheopenocean,corroboratingitsadaptiveroleinalgae-richhabitats.Exceptionalisthetoler- anceofLw-13eTagainstthebroad-spectrumantibiotictropodithieticacid,producedbyPhaeobacterspp.
co-occurringincoastalhabitats.Furthermore,Lw-13eTexhibitsfeaturesresemblingterrestrialplant- bacteriaassociations,i.e.biosynthesisofsiderophores,terpenesandvolatiles,whichmaycontribute tomutualbacteria-algaeinteractions.ClosestdescribedrelativeofLw-13eTisPseudopuniceibacterium sediminisCY03Twith98.4%16SrRNAgenesequencesimilarity.However,proteinsequence-basedcore genomephylogenyandaveragenucleotideidentityindicateaffiliationofLw-13eTwiththegenusPseu- dooceanicola.Basedonphylogenetic,physiologicaland(chemo)taxonomicdistinctions,weproposestrain Lw-13eT(=DSM29013T=LMG30557T)asanovelspecieswiththenamePseudooceanicolaalgae.
©2020TheAuthor(s).PublishedbyElsevierGmbH.ThisisanopenaccessarticleundertheCC BY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
The genus Pseudooceanicola of the alphaproteobacterial Roseobacter group (Rhodobacteraceae) currently comprises ten validated species of aerobic or facultatively anaerobic Gram- negativerods.Allso-fardescribedPseudooceanicolaspp.originate
∗Correspondingauthorat:NationalInstituteofAdvancedIndustrialScienceand Technology,Central6,Higashi1-1-1,Tsukuba,305-8566,Japan.
∗∗ Correspondingauthorat:InstituteforChemistryandBiologyoftheMarine Environment,Carl-von-Ossietzky-Str.9-11,26129Oldenburg,Germany.
E-mailaddresses:laura.wolter@uni-oldenburg.de(L.A.Wolter), t.brinkhoff@icbm.de(T.Brinkhoff).
fromseawater, deep-sea sediment or invertebrates. We herein describe the first Pseudooceanicola representative, designated Lw-13eT,fromthesurfaceofamarinemacroalga.StrainLw-13eT hasbeen isolated from Fucus spiralis, a brown macroalga with broaddistribution intidal areasalong the Europeanand North American Atlantic coast. Rhodobacteraceae constitute almost a quarteroftheepibacterialcommunityonFucusspp.[58,76]and physiologicalpropertiesofRhodobacteraceaeobtainedfromFucus surfacesindicateadaptationstoanepiphyticlifestyle[26].
Organismsinhabitingtidalflatsencounterregulardesiccation andrewettingeventsthatrequiredistinctadaptationstoswiftly changing environmental conditions suchas salt, oxidative and heatstress.Bacteriaassociatedwithcoastalmacroalgaearepoten-
https://doi.org/10.1016/j.syapm.2020.126166
0723-2020/©2020TheAuthor(s).PublishedbyElsevierGmbH.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc- nd/4.0/).
tiallyenrichedinadaptivefeatureslikeproductionofcompatible solutestodealwithosmoticstress[91]aswellashightolerance toantibioticsandheavymetals[83]thatcanaccumulateinalgal biomassfromterrestrialinput[33].Furthermore,thelifestyleof host-associatedbacteriaislikelyshapedbybiologicalinteractions, includingmechanismsofcommensalism,mutualismorparasitism betweenbacteriaandalgae[28].
Onerelevantaspectofbacteria-algaeinteractionsistheutiliza- tionofalgalconstituentsbybacterialepibionts,withspecialization for high- or low-molecular-weight compounds [35]. Moreover, bacteria-algae interactions can rely on chemical communica- tionandsecondarymetabolites,e.g.viathebacterialproduction of siderophores to access insoluble Fe3+ [72] and vitamins for auxotrophic algae [20], known for Rhodobacteraceae and their microalgalhosts[1].Stimulatorymechanismsincludethepromo- tionof algalgrowthbybacterialphytohormones[69],however, bothbacteriaandalgaecanalsoexertinhibitorymechanisms,e.g.
throughantifoulingcompoundsshapingtheepibioticcommunity [64].Productionofiron-scavengingmoleculesandtoxinsaswell asantibioticresistanceswereidentifiedasrelevantforbothmutu- alisticandantagonisticrelationshipsbetweenbacteriaandalgae [13].
Here, we identified traits in the Fucus-derived strain Pseu- dooceanicolaLw-13eTthatadaptforlifeonacoastalmacroalgae.
We expanded preliminaryinsightsonphysiologicaladaptations ofFucusepibionts[26]bydetailedinvestigationofgenomicand physiological featuresof strainLw-13eT.Its specificadaptations tolifeonmacroalgaein tidalareas,partiallyresemblingterres- trialplant-bacteriainteractions,distinguishstrainLw-13eT from seawater-, sediment-and invertebrate-derived Pseudooceanicola spp. Together with detection of a closely related phylotype in Fucus epibiota [26] and alginate lyase homologs in algae-rich coastalhabitats,ourstudyrevealsarangeofspecificadaptationsof strainLw-13eT.Supportedby(chemo)taxonomicdistinctionsand genomicanalyses,weproposestrainLw-13eTasanovelspecies withinthegenusPseudooceanicola.
Materialsandmethods
Samplecollectionandbacterialisolation
SpecimensofthebrownmacroalgaFucusspiraliswerecollected attheGermanNorthSeacoastinNeuharlingersiel(53◦4217.0N 7◦4216.1E)onJune27th2013duringlowtide.Algalspecimens weretransportedtothelabinacontainerat4◦Cwithintwohours andwashedthreetimeswithsterileseawatertoremoveloosely attachedbacteria.Subsequently,surfacesofalgalreceptacleswere swept overagarplates withMarineBroth Difco2216prepared withslightmodifications (hereafterreferred toasMB)toavoid precipitationofmediumcomponents[7]:12.6gMgCl2*6H2Oand 2.38gCaCl2*2H2OL-1andforthetraceelementsolution7mgNa- silicate*5H2Oand21.2mgboricacidL-1.Plateswereincubatedat 20◦Cinthedarkforthreedaysandsinglecoloniesre-streakedfour timesonfreshplatesforpurification,resultingintheisolationof strainLw-13eT[26].
Morphologicalandphysiologicalcharacterization
Cell morphology and motility were examined by light microscopy(AxioLabA1;Zeiss,Germany)inexponentialandsta- tionaryphase,bothinMBandartificialseawater(ASW)medium [90]supplementedwith3%MB.Thesupplementationwasfound tobeobligatoryforgrowthofstrainLw-13eTinASW(hereafter referred toasASW+MB).Cell motility and potentialtriggering byexogenous substanceswasinvestigatedinASW+MB supple-
mented with10mM sodium acetate, glucose, proline, maltose, N-acetylglucosamine, mannose,arabinose,fructose ordimethyl- sulfoniopropionate(DMSP),aswellaswith0.1%(w/v)polymeric and oligomeric alginate, oligomeric -d-mannuronate or Fucus powder[driedandshreddedalgalmaterial],respectively.Fortrans- mission electron microscopy, 50L of a culture grown in MB wereplacedonacoppergrid(200mesh;Plano,Germany),nega- tivelystainedusinguranylacetate,andanalyzedwithanEM902A electronmicroscope(Zeiss,Germany).Gram-staining,cytochrome oxidase and catalase activity as well as production of bacteri- ochlorophyllawereassayedasdescribedelsewhere[43].Analyses ofrespiratoryquinonesandcellularfattyacidswerecarriedoutby theGermanCollectionofCellCulturesandMicroorganisms(DSMZ, Braunschweig,Germany)(seeSupplementaryMethodsfordetails).
Growthexperiments
Testswithliquidcultureswereperformedintriplicatesintest tubeseachcontaining5mLmedium,shakenat150rpm.Eachtube wasinoculatedtoastartingOD600of0.001withcellsfromapre- culturegrownfor48hinMB.Unlessstatedotherwise,allgrowth experimentswerecarriedout at20◦Cin thedark.Growthwas followeddailybyOD600measurements,includingmediaandsub- stratecontrols.RangeforgrowthatdifferentpHvalueswastested betweenpH4and10.5,inincrementsof0.5anddeterminedin ASW+MBwith5mMproline.ThepHwasadjustedto4–8with 1MNaOHandwithglycineNaOHto8.5–10.5,followedbysterile- filtration.SalinitytolerancewastestedinNaCl-freeASW+MBwith 5mMproline,adjustedto0,0.5,1–10(in1%increments),12.5,15, 17.5and20%NaClusingsterile30%NaClsolution.Beforeinocu- lation,cellsfromthepre-culturewerewashedtwiceinNaCl-free ASW.TemperaturerangewasanalyzedinMBat4,7,9,15,20,24, 26,28,30,34,36and40◦C.Maximumgrowthrate(max)anddou- blingtime(td=ln2/max)weredeterminedunderoptimalgrowth conditions:inoculationtoastartingOD600of0.001in100mLMB, incubatedin500mLbaffledErlenmeyerflasksat28◦C,pH7.6and 150rpminthedark.Growthrateanddoublingtimeweredeter- minedbasedonOD600measurementseverytwohours,usinglinear regressionofasemi-logarithmicplotofmeanopticaldensity(from threereplicates)versustime.
Utilizationofdifferentcarbonsources(dissolvedinwaterand sterile-filtered)wasdeterminedwithfinalconcentrationsof0.1%
(w/v)foralginatesubstrates(polymericandoligomericalginate, oligomeric -d-mannuronate and Fucus powder) or 10mM for sugars(glucose,d-mannitol,d-mannose,N-acetylglucosamine,d- maltose,sucrose,fructose,l-arabinose)andaminoacids(alanine, proline,serine,valine,lysine,methionine,arginine,aspartate)after fivedays of incubation. The pre-culture wasgrown for 48h in MBandwashedtwiceinASWpriortoinoculation.Cellsgrownin ASW+MBwithoutfurtheradditionofcarbonsourceservedasneg- ativecontrol.Growthwasscoredasnegativewhenequaltoorless thanthenegativecontrol,andaspositiveaftertwotransfersand repeatedgrowthinthesamemedium.Reductionofnitrateand nitritewastestedinanoxicASW+MBcontaining0.5g/Lresazurin [23](seeSupplementaryMethods).
Antibioticandheavymetalsusceptibility
Antibioticsusceptibilitywastestedintriplicatesusinganantibi- oticdiscassayonMBagarplates[11]withpenicillinG,tetracyclin, streptomycinsulfate,chloramphenicol,kanamycinsulfate,specti- nomycin,gentamicinandampicillin(finalconcentrations1mM) andthemarinebroad-spectrumantibiotictropodithieticacid(0.1, 0.3,0.5,0.6and1mM).Plateswereinspecteddailyforinhibition zones.Controlsincludedsolventsoftheantibiotics(wateror50%
ethanol)aswellasMB.Heavymetaltolerancewastestedinmod-
ifiedliquidandonsolidMBwith0.04,0.075and0.1mMCuCl2or 1mMofarsenate/arsenite(SupplementaryMethods).Testswere performedintriplicates,withplatesortubeswithoutheavymetals servingascontrols.
Productionofsecondarymetabolites
Production and excretion of hemolysins was tested using a plate-basedbloodhemolysistest.Acellculturewasgrownfor48h inMBat20◦Cand100rpm.Subsequently,50Lcellsuspension wasinoculatedintoapiercedwellinColumbiabloodagarplates (MerckMillipore,Germany,No.146559)andoccurrenceofayel- low,clearringaroundthewellwithintwoweekswasscoredas
-hemolysis.Productionofvolatileorganiccompoundsandacyl- homoserine lactones wasanalyzedby GC/MSof CLSA andXAD cultureextracts(seeSupplementaryMethods).
Chemotactictriggeringofmotility
Chemotaxiswastestedonsoft(0.25%)agarplateswith10%MB ascarbonsource.TenLofabacterialculturegrownfor48hinMB wereinoculatedononesideoftheplateandtenLofthetested substancesopposite.Swimmingofbacteriatowardsorawayfrom the substance was scoredas chemotactic response. Substances analyzed for chemotactic response were: Fucuspowder, 1M of sodium-acetate,glucose,proline,500MofN-acetylglucosamine, maltose,mannose,arabinose,fructose,DMSP,aB-vitaminsolution [4],1%ofpolymericalginateandpolymeric-d-mannuronate.Soft agarplatesinoculated withbacteriabut nosubstanceservedas controlofmotilitywithouttriggering.
Genomesequencingandfunctionalanalysis
GenomicDNAofLw-13eTwasisolatedfromaculturegrown in MBfor48husingtheinnuPREPDNAMinikit(AnalytikJena, Germany).Unlessstatedotherwise,allsubsequentstepswereper- formed according to the manufacturer’s instructions. Extracted DNAwasusedtogenerateIlluminapaired-endsequencinglibraries (2×300bp) with the Nextera XT sample preparation kit (Illu- mina,SanDiego,CA).Generatedlibrariesweresequencedwitha MiSeqinstrumentandsequencingkitv3with600cycles(Illumina, SanDiego,CA,USA).Forgenomeclosure,high-molecular-weight DNA was isolated with the MasterPure Complete DNA & RNA Purification Kit (Biozym, HessischOldendorf,Germany).Quality ofisolatedDNAwascheckedbyagarosegelelectrophoresisand validatedonanAgilentBioanalyzer2100usingtheHighSensitiv- ityDNA12000Kit(AgilentTechnologies,Waldbronn,Germany).
DNA concentration and purity were checked with a Nanodrop ND-1000(PeqLabErlangen,Germany),followedbyexactquantifi- cation usingthe Qubit® dsDNA HSAssayKit(Life Technologies GmbH, Darmstadt, Germany). For Nanopore sequencing 1.5g high-molecular-weightDNAwasusedtopreparethreeindepen- dentlibrariesusingtheLigationSequencingKit1D(SQK-LSK109) and the NativeBarcode Expansion Kit (EXP-NBD103).Sequenc- ingwasperformedfor72husinga MinIONdeviceMk1Banda SpotON FlowCell R9.4.1(Oxford Nanopore Technologies) using MinKNOW software v19.05.0for sequencing and Guppy v3.0.3 [81]fordemultiplexing,resultingin19,955reads.Unicyclerv0.4.7 [88] wasusedwithdefaultsettingstoperformahybridassem- bly,resultingina closedchromosome(3,732,046bp)andthree closedplasmidswithsizesof202,410bp,111,009bpand49,258 bprespectively.RepliconswerevalidatedusingBandagev2.1[87]
andthegenomewasannotatedwithProkkav1.13.3[68].Putative biosyntheticgeneclusterswerepredictedusingAntiSMASHv4.1.0 [9],genomicislandsusingIslandviewerv4[6],andcarbohydrate- active enzymes using dbCAN2 [92]. Genome visualization and
comparisonwithotherPseudooceanicolaspp.wasdoneusingBRIG [2]. Homologs of the unique PL15 alginate lyase (Psal36760) weresearchedin77metagenomesfromdifferentmacroalgaeand seagrassesfromdiversecoastal habitatsin IMG [16]as wellas 243prokaryoticmetagenomesfromtheTARAexpedition(http://
bioinfo.szn.it/tara-blast-server-help-page/),only consideringhits withminimum65%querycoverageand60%aminoacididentity.
Phylogeneticanalysisanduniquegenes
Phylogeneticanalysisbasedonthefull-length16SrRNAgene (retrievedfromthecompletegenome)aswellas873core-genes wasperformedincontextwithcloselyrelatedstrainsidentified byBLASTn(https://blast.ncbi.nlm.nih.gov/Blast.cgi).Thegenomeof Pseudooceanicolalipolyticus(PGTB00000000)wasexcludedfrom further analyses due to high fragmentation (442 contigs with a medium size of 11kb, including many truncated genes and assembly contigs with kmer < 5). Whole-genome phylogeny wasperformedon873single-copyorthologous genesequences retrievedusingOrthoFinderv2.3.3[29].16SrRNAgenesequences andsingle-copyorthologswerealignedusingMAFFTv7.427[55]
andconcatenated.Theconcatenatedalignmentwasautomatically curatedforblocksrepresentedby>50%sequenceinformationusing GBlocksv0.19[79]andtheresulting188,268aminoacidpositions wereback-translatedtonucleotidedataforbetteralignmentreso- lutionusingtheembosspackage[66].Theconcatenated,curated andback-translatedalignment isshown inSupplementarydata file1.Maximum-likelihoodphylogeniesof16SrRNAandortholog genealignmentswerecalculatedusingRAxMLv8.2.12[73]withthe GTRCATmodeland1000or100bootstrapreplicates,respectively, followedbytreevisualizationusingMEGA-Xv10.0.5[45].Ruege- riameonggeiservedasoutgroup.DigitalDNA-DNAhybridization (dDDH)wascalculatedbythegenome-to-genomedistancecalcu- latorGGDC2.1[53]andaveragenucleotideidentities(ANI)using FastANI[40].
Toidentifycore,accessoryanduniquegenes,thegenomesof Lw-13eT,relatedPseudooceanicolaspp.andPseudopuniceibacterium sediminisastheclosestrelative ofLw-13eT (Table1)wereana- lyzedusingOrthoFinder,witha30%aminoacididentitythreshold.
Accessoryand unique geneswere functionallyannotated using eggNOG-mapperv2[37]andmanuallycurated.Ofthe3421unique genesinthetenstrains,32%werefunctionallyannotatedtodefined KEGGcategories(TableS1).EnrichmentofspecificKEGGcategories inthegenomesofLw-13eTandotherPseudooceanicolaspp.was analyzedastheratioofthespecificKEGGcategorycomparedto alluniquefunctionallyannotatedgenesintherespectivestrain.
Geneannotationswerecheckedbysequencecomparisonagainst theUniProtKBdatabase[82].Analysisofhomologiesofsinglegenes orgeneclusterswasdoneusingGeneiousv11.0.2(BiomattersLtd., Auckland,NewZealand).WeusetheabbreviationsPo.forPseu- dooceanicola,Pp.forPseudopuniceibacteriumandPh.forPhaeobacter throughoutthetext.
Resultsanddiscussion
Generalphysiological,phylogeneticandgenomicfeaturesofstrain Lw-13eT
Isolationandmorphologicalcharacterization
StrainLw-13eTwasisolatedfromthereceptaclesurfaceofthe commonbrownmacroalgaFucusspiralis,collectedattheGerman NorthSeacoastinsummerwhenbrownalgaehavetheirhighest physiologicalactivityandmostintenseinteractionwithepibiotic microbes[26,27].Lw-13eTwasamongthefirstorganismsform- ingcoloniesonMBagarplates. Aftertwo days,coloniesonMB
Table1
GenomestatisticsofLw-13eTandrelatedPseudooceanicolausedforcomparativeanalysis,includingsourceofisolation,GenBankaccessionnumbers,G+Ccontentand genomesize(inMbp)aswellasthefractionofcore,accessoryanduniquegenes.*excludedfromthecomparativeanalysisduetoahighfragmentationoftheassembly;**
excludedfromthecomparativeanalysisbecauseitisonlydistantlyrelatedtoPseudooceanicolaonwhole-genomelevel(Fig.3B).
Organism Locustag Isolationsource Genomeassembly G+C Mbp Genes Core(%) Accessory
(%)
Unique(%)
Pseudooceanicolaalgae Lw-13eT
PSAL BrownmacroalgaFucus spiralis,
NorthSea,Germany
CP060436–CP060439 64.1 4.09 3776 31.3 57.3 11.4
Pseudooceanicola antarcticusAr-45T
CVM39 Seawater(50m), SouthernOcean
GCF002786285.1 66.1 4.30 3992 29.6 61.1 9.3
Pseudooceanicolaatlanticus 22II-S11gT
ATO9 Seawater,
AtlanticOcean
GCF000768315.1 64.1 4.94 4633 25.5 66.0 8.5
Pseudooceanicolabatsensis HTCC2597T
OB2597 Seawater,
WesternSargassoSea
GCF000152725.1 66.1 4.44 4138 28.5 63.1 8.4
Pseudooceanicolaflagellatus DY470T**
B8991 Seawater, PacificOcean
GCF900176485.1 60.1 4.41 4143
Pseudooceanicolalipolyticus 157T*
Seawater, PhilippineSea
GCF002786325.1 64.6 5.24 4826
Pseudooceanicolamarinus CECT7751T
PSM7751 Coastalseawater(4–5m), Taiwan
GCF900172385.1 66.8 4.53 4244 27.8 62.5 9.7
Pseudooceanicola nanhaiensisSS011B1-20T
Q344 Sediments(1100m),South ChinaSea
GCF000688295.1 67.9 4.66 4442 26.6 64.8 8.7
Pseudooceanicola nitratireducensJLT1210T
BMX99 BeibuGulf, SouthChinaSea
GCF900109195.1 64.2 4.06 3840 30.7 62.9 6.4
Pseudooceanicolaonchidii XY-99T
E4S58 Onchidiumsp.(seaslug), SouthChinaSea
GCF 004959925.1 65 3.67 3528 33.5 61.8 4.7
Pseudopunicei-bacterium sediminisCY03T
DL237 Sediment(25mwater depth),YellowSea,China
GCF003575965.1 63.7 4.45 4036 29.2 62.8 8.0
Pseudooceanicolapacificus 216PA321T
GLS40 Deep-seasediment (5788m), PacificOcean
GCF009789075.1 66.3 3.89 3526 33.5 56.7 9.9
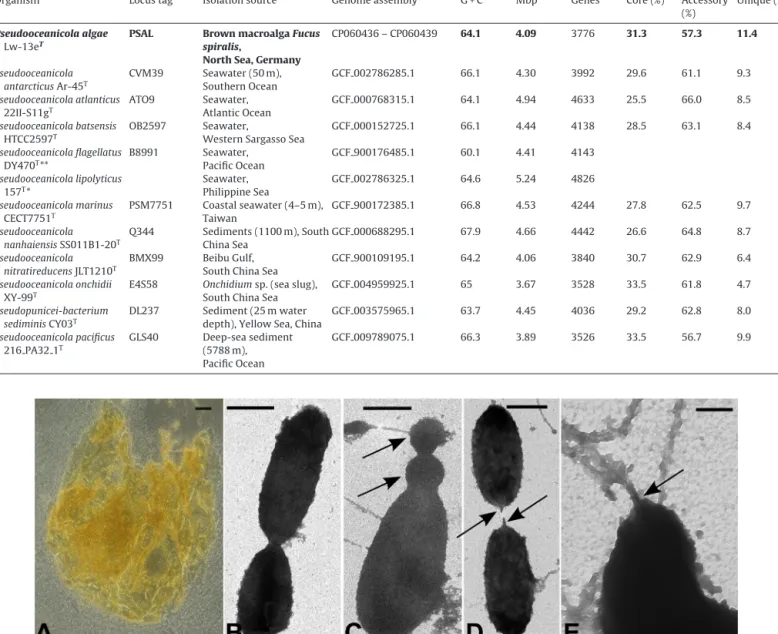
Fig.1. TransmissionelectronandlightmicroscopyofLw-13eT.DensecellaggregatesofLw-13eTarecharacterizedbyorangecoloration,potentiallythroughpigment production(A).Heterogeniccellmorphology,dividingviabinaryfission(B)andpotentialbudding(arrowsinC)andoftenconnectedbypilus-likestructures(arrowsinD).
Somecellsdisplayflagellum-likestructures(arrowinE)andseemedrepeatedlyconnected,butthesestructuresweredestroyedduringsamplepreparation(alsonotecell debrisinthesamples).Barsrepresent20m(A),1m(B,C,D)and0.2m(E).
appear cream-colored,circularandconvex,withashinysurface andadiameterofupto0.5mm.Afteroneweek,coloniesturnyel- lowishwithfuzzyedgesanddiametersupto3mm.Inliquidmedia (MBandASW+MB),Lw-13eTgrowscreamy-yellowish,withcells clumping in stickyhard-to-disruptaggregates, partlyappearing orangewhenconcentratedbycentrifugation(Fig.1A).Singlecells are irregular elongatedrods, 1.5–3mlong and approximately 1mwide,displayingheterogeneousmorphologiesinliquidcul- tures. Cells propagatethroughboth binaryfission (Fig.1B)and budding (arrowsin Fig. 1C), which might relate to CtrA-based phosphorelay(seebelow).Mostcellswereconnectedbypilus-like structuresasobservedbylightmicroscopy,whichwereprobably disruptedduringpreparationfortransmissionelectronmicroscopy (arrowsinFig.1D).Detectionofseveralflagellum-likeappendages (Fig.1E)correspondstothepresenceofflagella-encodinggenesin thechromosome(Fig.2,TableS2)withhomologytothefla1gene
clusterofDinoroseobactershibae(Fig.S1).However,cellswerenon- motile,independentofculturemediaorgrowthphase,andmotility wastriggeredneitherbyadditionofalgalmaterialoralgal-derived substancestothemedium.ThegenusPseudooceanicolawasprevi- ouslydescribedasnonmotile,althoughpolarorsubpolarflagella weresporadicallyreported[38,94]andPseudooceanicolagenomes harbor flagella-encodinggenes, withmotility indicated for two strains[5].TheprevalenceofflagellagenesingenomesofPseu- dooceanicolaspp.mightthusbeacommonfeature,butitremains tobedeterminedifthoseareexpressedonlyunderyetunknown conditionsorusedfor purposesotherthanmotility,e.g.surface attachment[71]. Discriminativegrowthcharacteristicsof strain Lw-13eTtorelatedstrainsaresummarizedinTable2.Acomplete overviewofgeneralgrowthparametersis showninthespecies description(Table3).
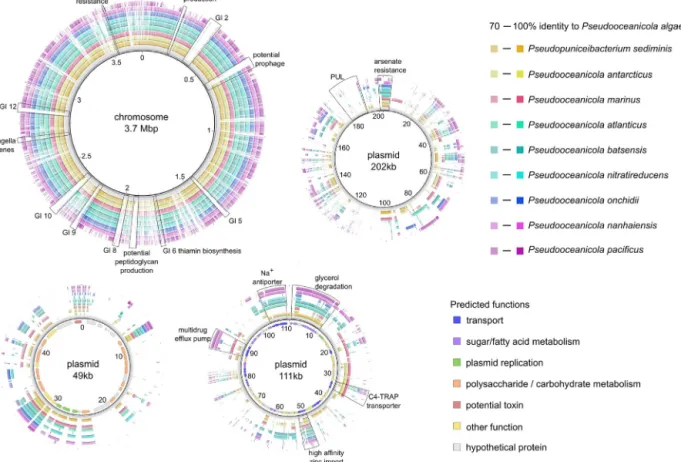
Fig.2. GraphicalrepresentationofthecompletegenomeofP.algaeLw-13eT.ColoredcirclesshowblastncomparisontootherPseudooceanicolastrainsasdescribedinthe legend,with%identityillustratedbycolorgradients.Functionalannotationsofthegenesonsmallerplasmids(49kb,111kb)aredepictedintheinnercirclebycolorationas giveninthelegend.Genomicislands(GIs)predictedusingIslandViewerv4(TableS3)areindicated.
Table2
DifferentialcharacteristicsofstrainLw-13eTcomparedtorelatedtypestrains.1:Lw-13eT;2:Pp.sediminisCY03T;3:Po.antarcticusAr-45T;4:Po.marinusAZO-CT;5:
Po.atlanticus22II-S11gT(typespecies).+,positive;−,negative;w,weak;n.d.,notdetermined;r,resistant;s,sensitive.Concentrationsoftestedsubstancesaregivenin thematerialsandmethodssection;*resultsarebasedonvalidlypublishedBiologdata.Althoughthisonlyallowsanindirectcomparisonofsubstrateutilization,our complementaryresultsprovideconclusiveevidenceofecologicaldifferentiationinlinewithspeciesseparation;**CompletefattyacidcompositionsareshowninTableS8.
Resultsfortheothertypestrainswereobtainedfromthespeciesdescriptions[38,48,49,93].GeneralcharacteristicsofstrainLw-13eTsharedwithotherPseudooceanicola spp.areshowninTable3.
Characteristic 1 2 3 4 5
Isolationsource surfaceofFucus spiralis
Marinesediment (25m)
Seawater(50m) Seawater(4–5m) surfaceseawater
Cellsize(m) 1×1.5–3 0.8–1.3×1.4–2.2 0.6×1 0.5×1 1×2.5
DNAG+Ccontent(mol%) 64.1 62.8 62 70.9 64.1
Colonycolor yellowcream opaque cream creamwhite faintyellow
Temperaturerange(◦C) 4–34 5–40 4–40 4–42 10–41
Temperatureoptimum(◦C) 20–28 30 35–37 28–35 25–28
Salinityrange(%NaCl) 0.5–17.5 0.5–9 0.5–10 2–8 0.5–9
Salinityoptimum(%NaCl) 0.5–7.5 1.5–2 0.5–3 3–5 1–7
Substratesused:
Alanine + n.d. −* + n.d.
d-mannose + + +* − −*
N-acetyl-glucosamine + n.d. +* − −*
Hydrolysisof:
Tween80 − − +* − −
Antibioticsusceptibility:
Ampicillin s s s r s
Gentamicin w n.d. s s s
Kanamycin r n.d. s s s
PenicillinG s s s r s
Streptomycin r n.d. s s s
Majorfattyacids(>10%)(in orderofabundance)**
C18:16c/7c(84%) C19:0cyclo8c(35%), C16:0(28%), C18:16c/7c(17%)
C16:0(34%), C19:0cyclo8c(33%), C18:16c/7c(21%)
C18:16c/7c(49%), C19:0cyclo8c(25%), C16:0(15%)
C18:16c/7c(55%), C16:0(16%), 11-methylC18:17c
(11%)
Table3
SpeciesdescriptionofstrainLw-13eTfollowingthedigitalprotologuestandard.
Speciesname Pseudooceanicolaalgae
Specificepithet algae
Speciesstatus sp.nov.
Speciesetymology al’gae,L.gen.n.algae,ofanalga,seaweed;referringtotheisolationsourcefromalgae Descriptionofthenewtaxonanddiagnostictraits Cellsarepleomorphicandnon-motile,despitegenomicandmicroscopicevidenceof
flagella.Singlecellsareirregularlyelongatedrods,1.5–3mlongandapproximately 1mwide,displayingheterogeneousmorphologiesinliquidcultures.Cellspropagate throughbinaryfissionandbudding.Coloniesareshinycream-colored,circularand convex,withadiameterofupto0.5mm.Afteroneweek,coloniesturnyellowishwith fuzzyedgesanddiametersupto3mm.Inliquidmedium,Lw-13eTgrows
creamy-yellowish,withcellsclumpinginstickyaggregatesthatarehardtodisrupt andappearorangewhenconcentratedbycentrifugation.Growthoccursat4–34◦C (optimum20–28◦C),atpH5.5–9.0(optimum6.5–8.0)andwith0.5–17.5%(optimum 0.5–7.5)NaCl.Underoptimalconditions,themaximalspecificgrowthrate(max)is 0.062h1,withadoublingtimeof11.2h(Fig.S2D).Testsforcatalaseandoxidaseare positive,reductionofnitrateandnitritearenegative.Thesolerespiratoryquinoneis ubiquinone-10,widelydistributedinAlphaproteobacteria.StrainLw-13eTdoesnot hydrolyzegelatin,starchorTween80.Itissusceptibletoampicillin,chloramphenicol, penicillinGandweaklytogentamicin,butisresistanttostreptomycinandkanamycin aswellastothebroadrangeantibiotictropodithieticacid(TDA).StrainLw-13eT growsonglucose,d-mannitol,alanine,d-mannose,N-acetyl-glucosamine,proline, d-maltose,d-fructose,sucroseandweakonserineandL-arabinose.Productionof iron-chelatingsiderophores,vitaminB12,terpenes,andvolatileswasdetected.A peculiarfeatureisthepredominanceofC18:17candC18:16cfattyacids(84%), contrastingtoagreaterdiversityoffattyacidsinotherPseudooceanicolaspp.
Countryoforigin Germany
Regionoforigin Neuharlingersiel,NorthSeacoast
Dateofisolation 27/06/2013
Sourceofisolation SurfaceofthemarinemacroalgaFucusspiralis
Samplingdate 27/06/2013
Latitude 53◦4217.0N
Longitude 7◦4216.1E
16SrRNAgeneaccessionnumber KM268063
Genomeaccessionnumbers CP060436–CP060439
Genomestatus complete
Genomesize 4,094,723
GCmol% 64.1
Numberofstrainsinstudy 1
DesignationoftheTypeStrain Lw-13e
StrainCollectionnumbers DSM29013=LMG30557
Miscellaneous,extraordinaryfeaturesrelevantforthedescription productionoforangepigment;heterogenicshapeofcells,propagatingthroughbinary fissionandbudding;non-motile,despitegenomicandmicroscopicevidenceofflagella
Genomicandphylogeneticanalysis
The closed genome of Lw-13eT consists of a chromosome (3732kb)andthreeplasmids (202,111and49kb)(Fig.2),with an overall G+C content of 64.1 %. The genome contains three rRNAgeneclusters,52tRNAgenes,onetmRNAgene,2951genes encodingproteinswithpredictedfunctions,and826genesencod- inghypotheticalproteins.Furthermore,18genomicislandswere predicted(TableS3),demonstratingmultiplepriorgenetictransfer events.
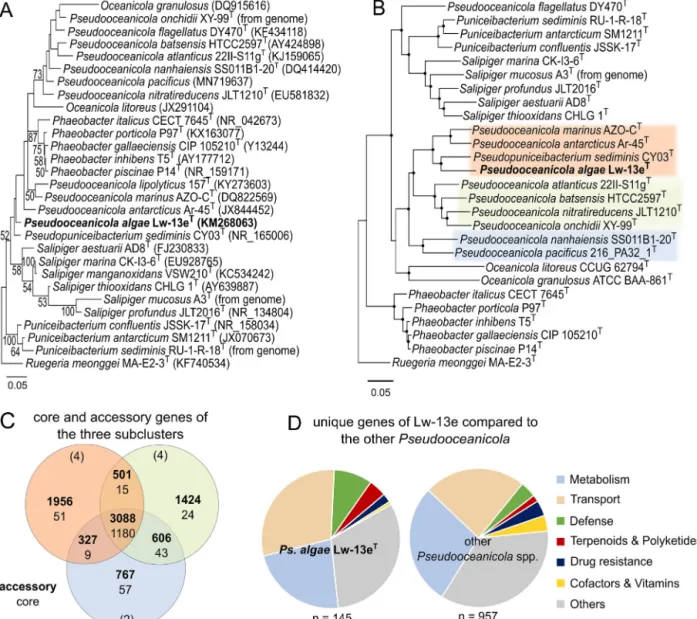
Phylogenybasedon16SrRNAgenesdemonstratedthatLw-13eT fallsbetweenitsclosestdescribedrelativePseudopuniceibacterium sediminisCY03T (98.4%16SrRNAgenesequence similarity)and aclustercontainingPseudooceanicola,OceanicolaandPhaeobacter species(Fig.3A).However,protein-basedcoregenomephylogeny shows clearaffiliation of Lw-13eT withthegenusPseudoocean- icola,forming asubcluster withPo.antarcticus,Po. marinusand Pp. sediminis (Fig. 3B, orange coloration) whereas other Pseu- dooceanicola spp. form two separate clusters (green and blue coloration).Po.flagellatuswasexcludedfromcomparativeanaly- ses,asitclusterswithSalipigerspp.andPuniceibacteriumspp.in whole-genomephylogeny.Deepbranchingfrombothgenerasug- gestsfuture reclassificationofPo.flagellatus.Averagenucleotide identities (ANI) of 79.6–79.9% with Pp. sediminis, Po. marinus and Po. antarcticus (Table S4) and <25% similarities using digi- talDNA-DNAhybridization(TableS5)illustratethatLw-13eTisa distinctspeciesinaccordancewithacceptedthresholdsof≤83%
ANI [40],and suggeststaxonomic reclassificationof Pp. sedimi-
nisasanotherPseudooceanicolaspecies.ThereclassificationofPo.
flagellatusandPp.sediminisis,however,beyondthescopeofthis study.
GenomeanalysisofLw-13eTandrelatedspecies
WeperformedacomprehensivegenomicanalysisforstrainLw- 13eTandrelatedPseudooceanicolaspp.(Table1)toidentifyfeatures thatmayreflectadaptationstothetidalhabitatandalgalassocia- tionofLw-13eT.The1180coregenesofallPseudooceanicolaspp.
constitute26–34%ofprotein-codinggeneswithinallgenomes, whereas 57–66%of identified orthologousgenesare accessory (Table1).ThethreePseudooceanicolasubclustersformingbycore- genomephylogeny(Fig.3B;orange,greenandbluecoloration)had fewcluster-specificgenes,i.e.phylogeneticrelatednessdoesnot correspondtofunctionalsimilarity(Fig.3C).Thisobservationsug- gestsaversatilelifestyleanddynamicadaptabilityofeachstrain, drivenbyspecificevolutionarypressurerelatedtotheirparticu- larenvironmentalorigin(Table1).Acrossallstrains,themajority ofuniquegeneswasaffiliatedwithKEGGcategories‘Transport’
and‘Metabolism’(Fig.3D),providingfurtherevidencethatmem- bersoftheRoseobactergroupencodea considerablediversityof transportersmediatingstrain-specificsubstrateadaptations[12].
UniquegenesofstrainLw-13eT(i.e.genesnotdetectedinany otherstrainwith>30%aminoacididentity)constituted11.4%ofall genes,beingthehighestfractionofallPseudooceanicolaspp.ana- lyzed(Table1).ComparedtorelatedPseudooceanicolaspp.,unique genes of Lw-13eT are enriched in functions related to defense
Fig.3. PhylogeneticplacementofstrainLw-13eTandrelatedstrainswithRuegeriameonggeiMA-E2-3Tasoutgroup,andKEGGcategorizationofuniquegenesofLw-13eT comparedtorelatedbacteria.(A)Maximum-Likelihoodphylogenycalculatedon16SrRNAgenesequenceswith1312nucleotidepositions.Bootstrapvalues>50%areshown basedon1000replications.Bar:0.05substitutionspernucleotideposition.(B)Maximum-Likelihoodphylogenybasedonproteinsequencesof873coregenes(overall188,268 aminoacidpositions).Nodeswithdotsaresupportedby100bootstrapreplicates.ColoredgroupsreflectthethreesubclustersofPseudooceanicolaforwhichfractionsofcore andaccessorygenesareshownin(C).Bar:0.05substitutionsperaminoacidposition.(C)Venndiagramshowingthedistributionofaccessory(bold)andcoregenesofthe threePseudooceanicolasubclusters(seeFig.3B).Numberofgenomeswithineachsubclusteraregiveninparentheses.(D)Fractionofuniquegenesassociatedwithdifferent KEGGcategoriesforstrainLw-13eTandrelatedstrains.KEGGcategoriesareshownasfractionsofuniquegeneswithdefinedfunctionalKEGGannotation(32%ofallunique genesretrievedbyOrthoFinder).Thecategory‘Metabolism’includes‘Aminoacid,Carbohydrate,Energy,LipidandNucleotidemetabolism’,‘Metabolism’and‘Metabolism ofotheraminoacids’.Thecategory‘Transport’includes‘Membranetransport’and‘TransportandCatabolism’.(Forinterpretationofthereferencestocolourinthetext,the readerisreferredtothewebversionofthisarticle.)
strategies(salt,heatandoxidativestressaswellasrestrictionmod- ificationsystemsforphagedefense;Fig.3D,greencoloration)anda uniqueabilityfortheproductionofterpenoids(redcoloration).In contrast,uniquegenesofotherPseudooceanicolaspp.areenriched in theproduction of specificdrugresistancemechanisms(dark blue)andcofactorsandvitamins(yellow).Nevertheless,strainLw- 13eTdoesencodeanaccessoryclusterforvitaminB12biosynthesis.
ThepreviouslyshownproductionofthisvitaminfortheB12aux- otrophF.spiralisunderlinestheassociationofLw-13eTtothisalga [26].
Habitat-relatedadaptations
Tolerancetochallengingenvironmentalconditions
Nearlyone thirdof theunique genesinLw-13eT encodefor membranetransportproteins(Fig.3D).AsetofuniqueABC-type
transporterspredictedfortransportofcompatiblesolutesinclud- ingglycine/betaineandtaurine(TableS2)potentiallyenablesstrain Lw-13eTtowithstandtheosmoticstressintidalhabitats,supported bygrowthinasalinityrangeof0.5–17.5%(Table2).Thisbroadsalin- ityrangeofepibioticcommunitymembersmightlikewiseenable acclimationofbrownalgaewhenencounteringfreshwaterinflow [25].Otheruniquetransportergenesfromthegroupofmultidrug exportersenableLw-13eTtotolerateawiderrangeofantibiotics thanrelatedPseudooceanicolaspp.(Table2).Furthermore,strain Lw-13eTshowsconsiderabletoleranceagainstthebroad-spectrum marineantibiotictropodithieticacid(TDA),withoutgrowthlimi- tationuntil0.6mMofTDA(Fig.S2A).Thistoleranceisinthesame rangeasinTDA-producingPhaeobacterspp.[11],whereassensitive strainsarenormallyinhibitedby0.1mM[61].TolerancetoTDAby non-TDAproducingstrainsisrarelydescribedtodate[61]andpos- siblyalsorelatestotheabundanceofmultidrugexporters,asthe
proposedresistancegenestdaR1-R3fromPhaeobacterspp.[89]are absentinLw-13eT.TDA-producingPhaeobacterspp.arealsopreva- lentonsurfacesandintidalhabitats[11]andmighthenceco-occur withLw-13eT,stimulatingtheevolutionofresistance.Growthwith 1mMarsenate(Fig.S2B)andupto0.1mMcopper(Fig.S2C)cor- respondstothepresenceofrelatedresistancegenes(Fig.2,Table S2).Thecompletesetofcopperandarsenateresistancegenesare sharedwithPo.batsensis,isolatedfromtheSargassoSea(Table1) thatharborsvastamountsofseaweed[85].Thesetraitsmightthus bespecificforbacteriafromenvironmentswithhighabundanceof macroalgae,knowntobeenrichedinheavymetals[33].
Oligo-alginatedegradation
AspecificadaptationofstrainLw-13eT tomacroalgalassoci- ation is the presence of a unique alginate lyase gene encoded inapolysaccharideutilizationlocus(PUL)onthe202kbplasmid (Figs.2 &4A).Alginateisa linearpolysaccharidecomposedof
␣-l-guluronate(G)and-d-mannuronate(M)andamajorcom- ponentofthecellwallmatrixinbrownalgae,constituting∼50%
ofthedryweightofF.spiralis[51].ThealginatelyaseofLw-13eT is predicted asan exolytic oligo-alginate lyase fromthefamily PL15[50].Accordingly, strainLw-13eT grows onoligomericbut notonpolymericalginate(Fig.4B).Interestingly,theoligo-alginate lyaseof strainLw-13eTshows apreference formixedoligo-GM ratherthanmannuronate-richoligomers(oligo-M),comparedto thedegradationofalloligomericalginatetypesbyarelatedPL15 from the terrestrial plant-associated Agrobacterium fabrum C58 (Fig. 4C) [56]. The PUL also encodes kdgF and kdgK genes for downstream processingofalginatemonomersaswellasa pre- dictedfabGdehydrogenase/reductasegene,whichmightreplace dehR analogous toother alginolytic bacteria[39,44,78]. Besides homologytothePL15ofA.fabrumC58,wedetectedhomologous gene clustersin roseobacters isolatedfrom phytoplankton sur- facesorfromenvironmentswithhighconcentrations ofmacro- ormicroalgae(Fig.4C)[8,17,46,47].InMarinovumalgicolaDG898, thePULislocatedonachromiddevotedtocarbohydratetrans- portandbiosynthesis,enablingthestraintothriveincarbon-rich phycosphereenvironments[30].PresenceofthePULonthelinear chromosomeofA.fabrumC58suggeststhatthePULwasexchanged between theterrestrial and themarineenvironment, reflecting transfereventsofmarineplasmidsacrossbiogeographicandphy- logeneticbarriers[59].
Asthealginatelyaselacksasignalpeptide,apossiblescenario isthatoligomersaretakenupbythePUL-encodedputativesugar ABCtransporter(typeI)intotheperiplasmforexolyticcleavage.
Thedegradationofoligo-alginatesuggeststhatLw-13eTisasec- ondary consumeronalgalsurfaces,utilizing oligomersreleased byotherassociatesencodinglyasesspecificforcomplexalginate polymers.Suchcross-feedingonalgalcellwallconstituentsillus- tratesapartitioningofdifferenttaxaintopioneersandharvesters [3].ThepresenceofaPULinLw-13eTisnoteworthyasmembers of theRoseobactergrouparerarelydescribedasoligosaccharide degraders.AlgaeadaptationofLw-13eTisunderlinedbytheuseof variousotheralgalsubstrates,includingFucuspowderandproline (Fig.4B),mannose(Table2)andmannitol[26],allbeingenriched inbrownalgalbiomass[42].
Productionofvolatileorganiccompounds,terpenesand siderophores
Bacteriallyproducedvolatilecompoundsmediateinterspecies and interkingdom communication [67] as well as plant devel- opment anddefense in terrestrialhabitats [41]. StrainLw-13eT produces numerous volatiles,includingdimethyldi- andtrisul- fides,acetoinderivatives,aromaticcompoundsincludingnitroge- nouscompoundssuchaspyrazines,aswellasaliphaticketones (Table S6&Fig. S3).Someof theseare alsoproducedby other
surface-associatedroseobacters[34,80],indicatingaconsiderable potentialfor chemical communication.Dimethyl di- and trisul- fidesareknowngrowthpromotersordefensemoleculesagainst reactiveoxygenspecies(ROS)fromterrestrialplants[14,54],sug- gesting thatproduction byLw-13eT mightmediate comparable defenses against ROS produced by macroalgae [86]. Of further interestarethearomaticcompounds2-aminoacetophenoneand phenol,not commonlydescribed forbacteriaoftheRoseobacter group.InPseudomonasaeruginosa,2-aminoacetophenoneregulates antibiotictoleranceviaquorumsensing[62],whereasphenolpro- ductionisknownfromentericandlacticacidbacteria[19].Thered algae-associatedPseudovibriosp.D323waspostulatedasprovider ofphenolic-baseddefensecompoundscommoninbrownalgae[95]
withe.g.fishdeterrenteffects[74].Thus,provisionofphenolbyLw- 13eTcouldbeanotheradaptivefactorformacroalgalassociation bystrengtheningalgaldefense.Detectionofsaturatedandunsatu- rateddo-andtridecanonesasmajorvolatilesmatchesthefrequent detectionofaliphaticketonesandalcoholsoriginatingfromfatty acidbiosynthesisinmarineandotherbacteria[24].
Detectionofthevolatileterpeneslimonene,nerolidolandfar- nesol (Table S6) is consistent with enrichment of genes from KEGG category ‘Terpenoid and Polyketide Synthesis’ (Fig. 3D) andunprecedentedforroseobacterstodate.Terpeneproduction of Lw-13eT mayhave ecological implications as feeding deter- rent,membranestabilizer,anti-oxidant,signalingorantagonistic molecule[32,60].Productionoflimonenecouldprovideantimi- crobial defense for the algal host [77], while farnesol inhibits quorumsensingofPseudomonasaeruginosa[22]andmighthence influence bacterial communication within macroalgal epibiota.
PolyketideproductioninRoseobactergroupbacteriawasindicated before[52]butrequiresfurtherinvestigation.Terpenemetabolism in Lw-13eT includes a unique biosynthetic gene cluster with 40% amino acid identity to a squalene-producing gene cluster inRhodopseudomonaspalustrisATCCBAA-98(Rhizobiales)(Fig.2
&TableS2).Thishomologyindicatespotentialtransferbetween habitats,comparabletothealginatelyasedescribedabove.Pro- duction of squalene/lycopene as precursors for hopanoids and carotenoids[57],knowntoinfluencecellwallrigidity[10],might explaintheorangepigmentationoftheLw-13eTcellpellet(Fig.1A).
Terpene production among Rhodobacteraceae was so far only indicated for a Tateyamariaisolate frombobtail squid [18] and anuncultured metagenome-assembledgenome associatedwith microalgae[63].Accordingly,in75analyzedgenomes,homologs oftheterpene-relatedgeneclusterwereonlydetectedinLimimari- colahongkongensisDSM17492fromacoastalseven-dayoldbiofilm (∼50%sequenceidentity;e-value<10−5).
Non-ribosomalpeptidesynthesisinLw-13eTcorrespondstoa siderophore-relatedcluster(TableS2)with>42%aminoacididen- titytoenterobactinsynthesisgenesinE.coliK12[21].Enterobactin isoneofthestrongestbacterialsiderophores[65],explainingthe previouslyshown highiron-chelating activityofstrainLw-13eT [26].Siderophoreproductionmaybebeneficialforthebacterium anditsalgalhostinthetypicallyiron-limitedmarineenvironment [72].
Importanceofadaptivetraitsfordistributionandecologyinsitu Wedetectedatleastonehomolog(≥65%coverageand≥60%
aminoacididentity)ofthePL15inapproximately75%ofepibac- terialmetagenomesderivedfromdiversecoastalmacroalgaeand seagrasses(TableS7).TheprimarydetectionofPL15homologson seagrasses,macroalgaeandthesurroundingseawatersupportsthe notionthatoligo-alginatedegradationisacommoncharacteristic ofalgaeassociationincoastalenvironments.Incontrast,homologs wereonlyretrievedinthreeof243open-oceanTARAmetagenomes (TableS7).Thelongestalignmentwiththehighestidentity(99%
coverageand65%identity)wasretrievedfromtheepibioticcom-
Fig.4.Uniquepolysaccharideutilizationlocus(PUL)instrainLw-13eTandgrowthonoligomericalginate.(A)Plasmid-encodedPULharboringPL15oligo-alginatelyase (green),atype-IsugarABCtransporter(blue)andgenesforalginatemonomerprocessing(orangeandgrey).FramedgenesareuniquetoLw-13eTcomparedtoother Pseudooceanicolaspp.(B)GrowthexperimentsdemonstratedthatLw-13eTdegradesmixedguluronate-mannuronateoligomers(Oligo-GM)butnotmannuronate-rich oligomers(Oligo-M)andpolymericalginate(Poly-A),supportingfunctionalityofthePUL.Inaddition,Lw-13eTwasabletogrowonFucuspowder.Prolinewasusedasa positivecontrolformacroalgal-derivedsubstrates.(C)Homologousclustersweredetectedinfewrelatedroseobactersisolatedfromphytoplanktonsurfacesorhabitatsrich inalgalbiomass,aswellastheterrestrialplant-associatedAgrobacteriumfabrum.ID,%identity;QC,%querycoverageofthegenealignment.AsthegenomeofOceanicola granulosusHTCC2516isadraftgenome,thelocationofthePULhomologeitheronthechromosomeoramobilegeneticelementcannotbespecified.(Forinterpretationof thereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
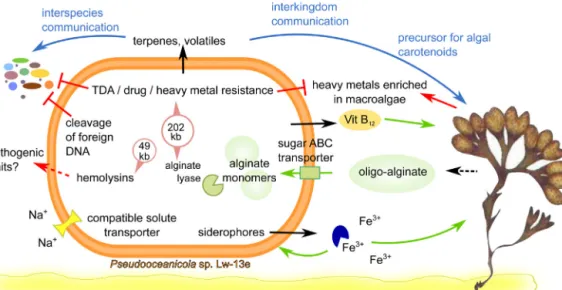
Fig.5. SummaryofadaptationsofLw-13eTtomacroalgaeassociationandtidalhabitats.Fromlowerleftcounterclockwise:Broadsalinitytolerancebasedontransporters forcompatiblesolutes(yellow);productionofiron-chelatingsiderophores(bluecircle);degradationofoligo-alginatebyPL15alginatelyase;productionandreleaseof vitaminB12;heavymetalresistance;interspeciesandinterkingdomcommunicationthroughproductionofterpenesandvolatiles;hemolysinproductionaspotentialmean ofpathogenicity.(Forinterpretationofthereferencestocolourinthisfigurelegend,thereaderisreferredtothewebversionofthisarticle.)
munityofthebrownalgaEckloniaradiata(BotanyBay,Australia), whichcontainsupto30%ofalginateandmannitol[75]andhence substratesaccessibleforLw-13eT.DetectionofPL15homologsin Australiaindicatesthatadaptivefeaturesenablegrowthofrelated Pseudooceanicolaspp.onalgalsurfacesworldwide.Furthermore, a16SrRNAgenephylotypewith99%sequencesimilaritytoLw- 13eTwasdetectedat0.02%relativeabundanceintheepibacterial community ofF.spiralisin theNorthSea[26],fromwhere Lw- 13eTwasisolated.Overall,theseenvironmentaldataillustratethat adaptive traits indeedenableniche occupation insitu,allowing thecolonizationofcoastalalgae-richhabitatsonwidegeographic scales.
Otherdistincttraits Potentialpathogenicity
Strain Lw-13eT has pathogenic potential by secreting membrane-destructinghemolysins,corroboratedby-hemolysis onblood agarplates (datanot shown). Thisobservationcorre- spondsto two unique genes coding for hemolysin production, onecontainedinatype-1secretionsystem(TableS2).Hemolysin production can drive virulence in macroalgal pathogens under elevatedtemperatures [31] and a comparable mechanism was demonstratedforPh.inhibens,changingfrommutualtopathogenic behaviorupon sensingof infochemicals[70].Thissuggeststhat