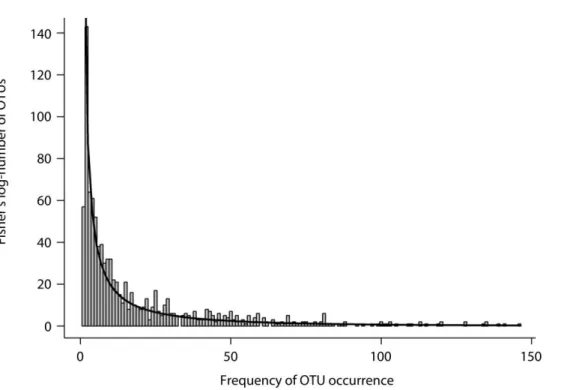
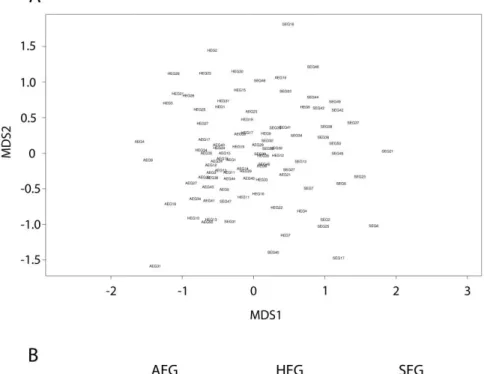
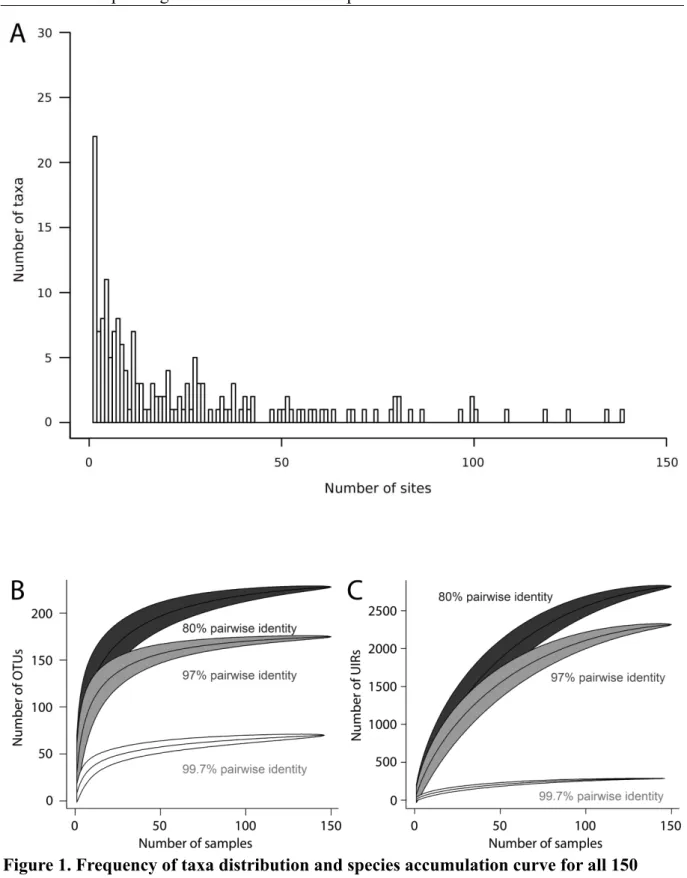
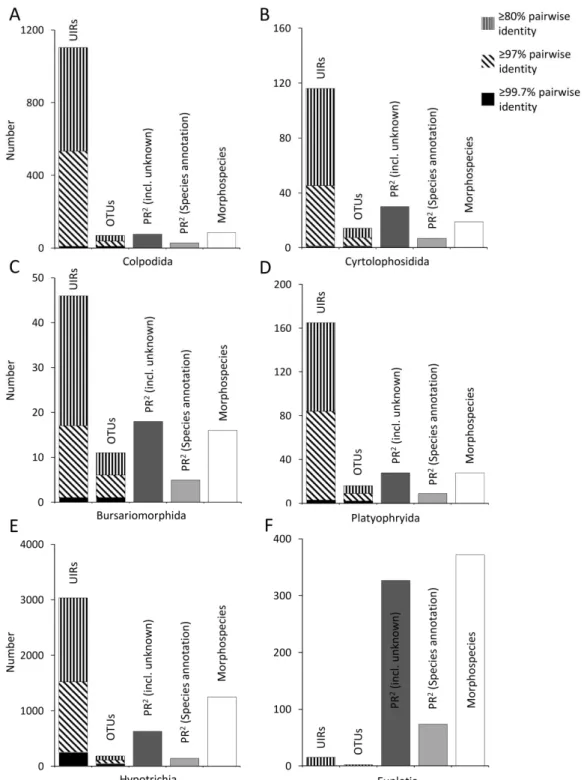
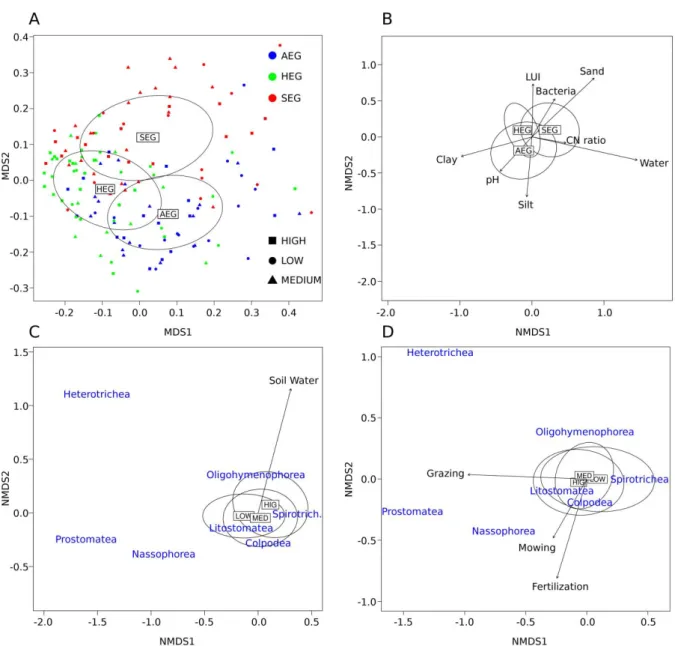
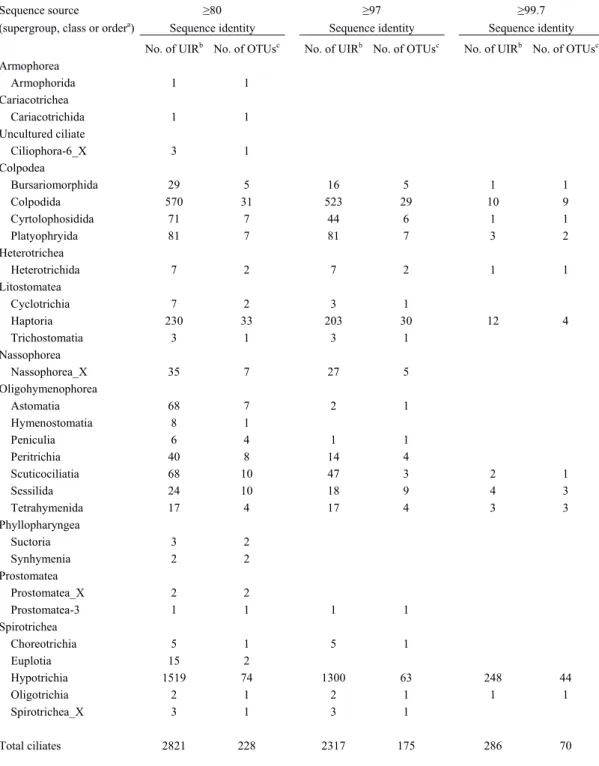
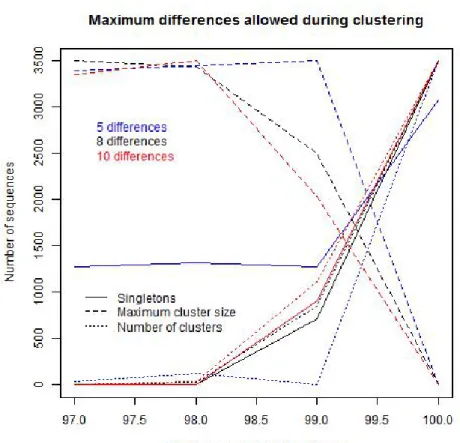
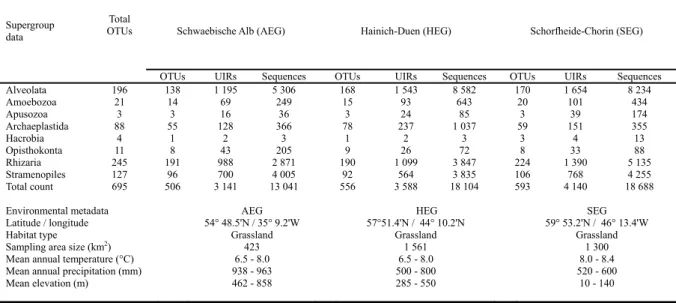
of German grassland soils
Volltext
Abbildung
ÄHNLICHE DOKUMENTE
We used an complementary approach based on light microscopy, HPLC-based phytoplankton pigment determination, in situ chlorophyll-a fl uorescence measurements, fl ow cytometry, molecular
Results I: Spatiotemporal variations influencing benthic bacterial communities.. Characterising sublittoral benthic bacterial communities. I: Spatiotemporal variations
Adjoint functors between module categories are described by a tensor and a Hom functor and the properties derived from the categorical setting are explained in Section 3.. Algebras
parapatric speciation model of Doebeli and Dieckmann (2003) by letting dispersal and
The regional and national summary listed in Table 6 and the class distribution shown in Figure 4 point to the fact that low yield and fair to good quality are the main
In disk storage operations, used to store and control disk sector address.. Retains address of low-order augend digit during addition for
In the last Section, we use his mass formula and obtain a list of all one- class genera of parahoric families in exceptional groups over number fields.. Then k is
properties of the substance, including C oxidation state and number of C atoms. In contrast, it was found that rates of LMWOS-C cycling within the microbial cells cannot be